Preclinical, Clinical, and Translational Sciences
(T1330-02-11) A PBPK Model for Acetaminophen: Incorporation of SULT Enzyme Ontogeny to Predict Age-Dependent Metabolism and Systemic Exposure in Pediatric Patient Populations
Tuesday, October 22, 2024
1:30 PM - 2:30 PM MT
- DT
David R. Taft, Ph.D.
Professor
Long Island University
Brooklyn, New York, United States - SS
Sonia Sharma
Ph.D. Student
Long Island University
Brooklyn, New York, United States
Presenting Author(s)
Main Author(s)
Purpose: To develop a PBPK model for acetaminophen in different pediatric patient populations that accounts for ontogeny of sulfotransferase (SULT) isozymes that play a critical role in acetaminophen metabolism.
Methods: PBPK modeling and simulation were performed using the Simcyp® Simulator (Version 21). The model incorporated the developmental ontogeny of four key hepatic SULT enzymes: SULT1A1, SULT1A3, SULT1B1, and SULT2A1. “Best-fit” ontogeny equations for each isozyme were determined by nonlinear regression analysis of enzyme abundance versus age (Table 1). A substrate profile for acetaminophen was created based on known physiochemical properties and pharmacokinetic parameters. The model was used to simulate the systemic exposure of acetaminophen after IV administration in various patient populations: Neonates (birth – 28 days), Infants (1 – 24 months), and Children (2-12 years). Model verification was based on visual predictive check (comparison of model-simulated plasma-concentration time profile compared with to observed clinical data1) and fold-error (ratio of model predicted and observed values) for steady state Cmax and AUC.
Results: SULT enzymes contribute significantly to acetaminophen metabolism in pediatric patients, most notably neonates (Table 2). The Simcyp®-simulated pharmacokinetic profiles for acetaminophen agreed with observed clinical data for systemic exposure (Cmax, AUC) (Table 3), and the model captured systemic levels over time in neonates, infants, and children.
Conclusion: The impact of SULT enzymes on drug metabolism is significant in neonates, and an important clinical consideration for acetaminophen. A PBPK model that incorporates SULT ontogeny has the potential to help guide dosing decisions in this vulnerable patient population.
Acknowledgements: The authors thank Dr. Bhagwat Prasad (Washington State University) for providing the SULT enzyme abundance data. Certara UK Limited (Simcyp Division) granted access to the Simcyp Simulator through a sponsored academic license (subject to conditions).
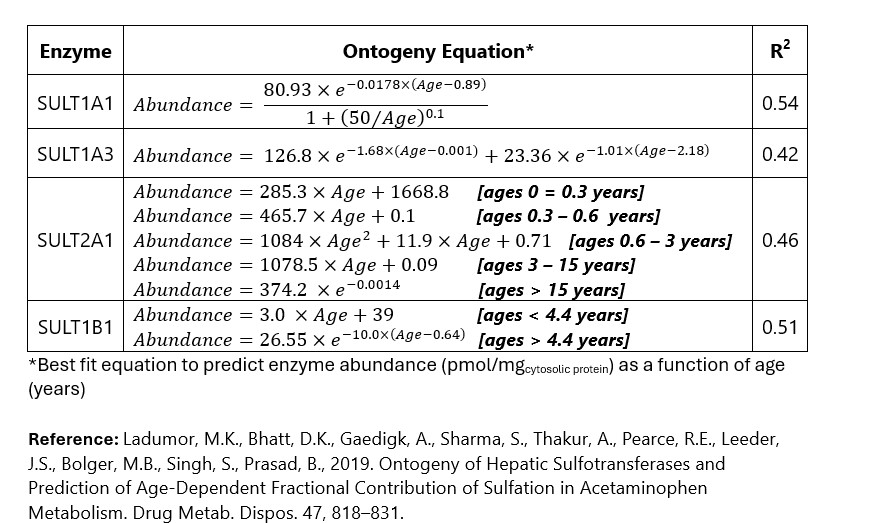
Table 1. Equations Used to Model SULT Ontogeny
.jpg)
Table 2. PBPK Model
Results: Mean % Contribution of SULT Isozymes to Acetaminophen Metabolism Across Pediatric Patient Groups
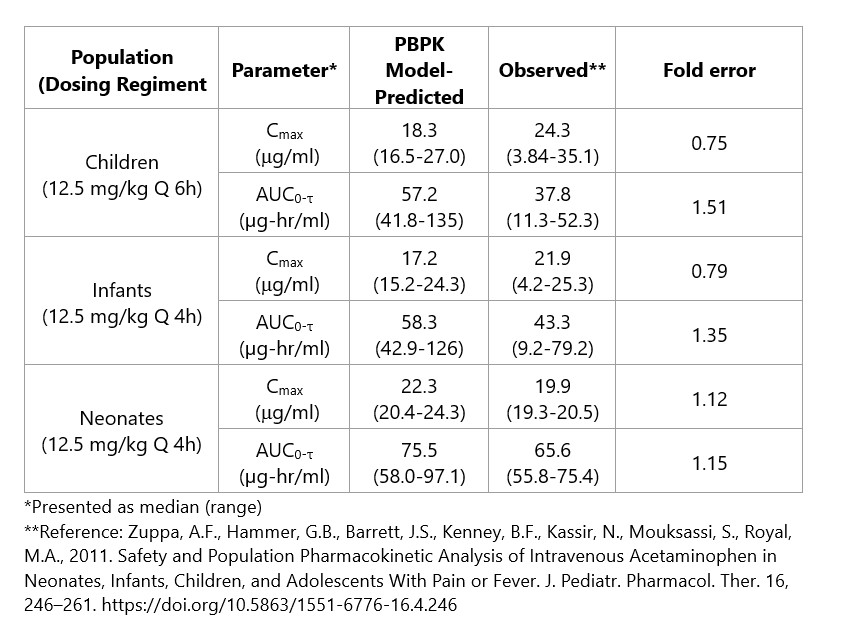
Table 3. Verification results for acetaminophen PBPK model using the Simcyp® Simulator.
Methods: PBPK modeling and simulation were performed using the Simcyp® Simulator (Version 21). The model incorporated the developmental ontogeny of four key hepatic SULT enzymes: SULT1A1, SULT1A3, SULT1B1, and SULT2A1. “Best-fit” ontogeny equations for each isozyme were determined by nonlinear regression analysis of enzyme abundance versus age (Table 1). A substrate profile for acetaminophen was created based on known physiochemical properties and pharmacokinetic parameters. The model was used to simulate the systemic exposure of acetaminophen after IV administration in various patient populations: Neonates (birth – 28 days), Infants (1 – 24 months), and Children (2-12 years). Model verification was based on visual predictive check (comparison of model-simulated plasma-concentration time profile compared with to observed clinical data1) and fold-error (ratio of model predicted and observed values) for steady state Cmax and AUC.
Results: SULT enzymes contribute significantly to acetaminophen metabolism in pediatric patients, most notably neonates (Table 2). The Simcyp®-simulated pharmacokinetic profiles for acetaminophen agreed with observed clinical data for systemic exposure (Cmax, AUC) (Table 3), and the model captured systemic levels over time in neonates, infants, and children.
Conclusion: The impact of SULT enzymes on drug metabolism is significant in neonates, and an important clinical consideration for acetaminophen. A PBPK model that incorporates SULT ontogeny has the potential to help guide dosing decisions in this vulnerable patient population.
Acknowledgements: The authors thank Dr. Bhagwat Prasad (Washington State University) for providing the SULT enzyme abundance data. Certara UK Limited (Simcyp Division) granted access to the Simcyp Simulator through a sponsored academic license (subject to conditions).

Table 1. Equations Used to Model SULT Ontogeny
.jpg)
Table 2. PBPK Model
Results: Mean % Contribution of SULT Isozymes to Acetaminophen Metabolism Across Pediatric Patient Groups

Table 3. Verification results for acetaminophen PBPK model using the Simcyp® Simulator.
