Discovery and Basic Research
(T1130-06-35) Leveraging Machine Learning to Predict Clinical Adverse Events Driven by Target-Specific and Off-Target Binding
Tuesday, October 22, 2024
11:30 AM - 12:30 PM MT
- LZ
Luchen Zhang, MS (he/him/his)
Graduate Student
University of Michigan
Ann Arbor, Michigan, United States - AC
Albert Cao
Student
University of Michigan
Ann Arbor, Michigan, United States - YB
Yingzi Bu, Ph.D.
Post-doc Fellow
University of Michigan
Ann Arbor, Michigan, United States - DS
Duxin Sun, Ph.D.
Professor
University of Michigan
Ann Arbor, Michigan, United States
Presenting Author(s)
Main Author(s)
Co-Author(s)
Purpose: Currently, 90% of clinical drug development fails, where 30% of these failures are due to clinical toxicity. The current extensive animal toxicity studies are not predictive of clinical adverse events (AEs) at clinical doses, while current computation models only consider very few factors with limited success in clinical toxicity prediction. We aimed to address these issues by developing a machine learning (ML) model that integrates drug interactions with both on- and off-targets and expression levels of these targets in various tissues for clinical AEs prediction.
Methods: A collection of 759 FDA-approved drugs with known AEs was adopted directly from Galeano et al.[1]. Protein target expression levels across various tissues were sourced from the Human Protein Atlas[2]. We first adapted the reported ConPLex ML model[3] to predict the drugs’ binding affinities (pIC50) for their intended targets and off-targets among 477 protein targets. Contrastive learning was incorporated to refine the accuracy of the predicted drug-target binding affinities. Subsequently, we developed a multi-layer perceptron (MLP) structure to predict clinical AEs incorporating these predicted drug-target interactions along with the expression profiles of on-/off-targets across various tissues. The model training involved a thorough split of data into training, validation, and testing sets with a ratio of 0.68:0.17:0.15 and utilized weighted binary cross-entropy to address label imbalance within the dataset. The AdamW optimizer (learning rate initialization of 10−5 and weight decay of 10−5) and early stopping techniques were leveraged during model training to optimize performance and mitigate overfitting, respectively.
Results: The utilized dataset of 759 FDA-approved drugs displayed a broad spectrum of structural diversity (Figure 1a), underscoring the versatility of our model in predicting AEs for compounds with a wide range of structural variations. However, traditional dimensionality reduction techniques (Figure 1b) did not effectively segregate compounds based on their associated AEs, emphasizing the multifaceted nature of drug behavior and the need for more advanced modeling approaches to capture these intricate relationships. The adapted ConPLex model predicted drug-target interactions for both on- and off-target effects, as shown by the correct co-localization of the six small molecule kinase inhibitors with their respective kinases (Figure 2). The further constructed MLP model demonstrated good predictive capability of clinical AEs such as headache, nausea, diarrhea, and dizziness, yielding high metrics for accuracy (exceeding 0.86), precision (exceeding 0.88), recall (exceeding 0.96), and F1-scores (exceeding 0.92) (Table 1).
Conclusion: This study presents an advanced machine learning model that integrates drug structure data, on- and off-target binding interactions, and tissue-specific target expression profiles for the prediction of clinical AEs. Our approach provides a new insight into the mechanistic understanding of in vivo drug toxicity in relationship with drug on-/off-target interactions. The coupled ML models, once validated with larger datasets, may offer advantages to directly predict clinical AEs using in vitro/ex vivo and preclinical data, which will help to reduce drug development failure due to clinical toxicity.
References: [1] Galeano D, Li S, Gerstein M, Paccanaro A. Predicting the frequencies of drug side effects. Nature Communications. 2020 Sep 11;11(1):4575.
[2] Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, Olsson I. Tissue-based map of the human proteome. Science. 2015 Jan 23;347(6220):1260419.
[3] Singh R, Sledzieski S, Bryson B, Cowen L, Berger B. Contrastive learning in protein language space predicts interactions between drugs and protein targets. Proceedings of the National Academy of Sciences. 2023 Jun 13;120(24):e2220778120.
Acknowledgements: The authors declare no conflict of interest.
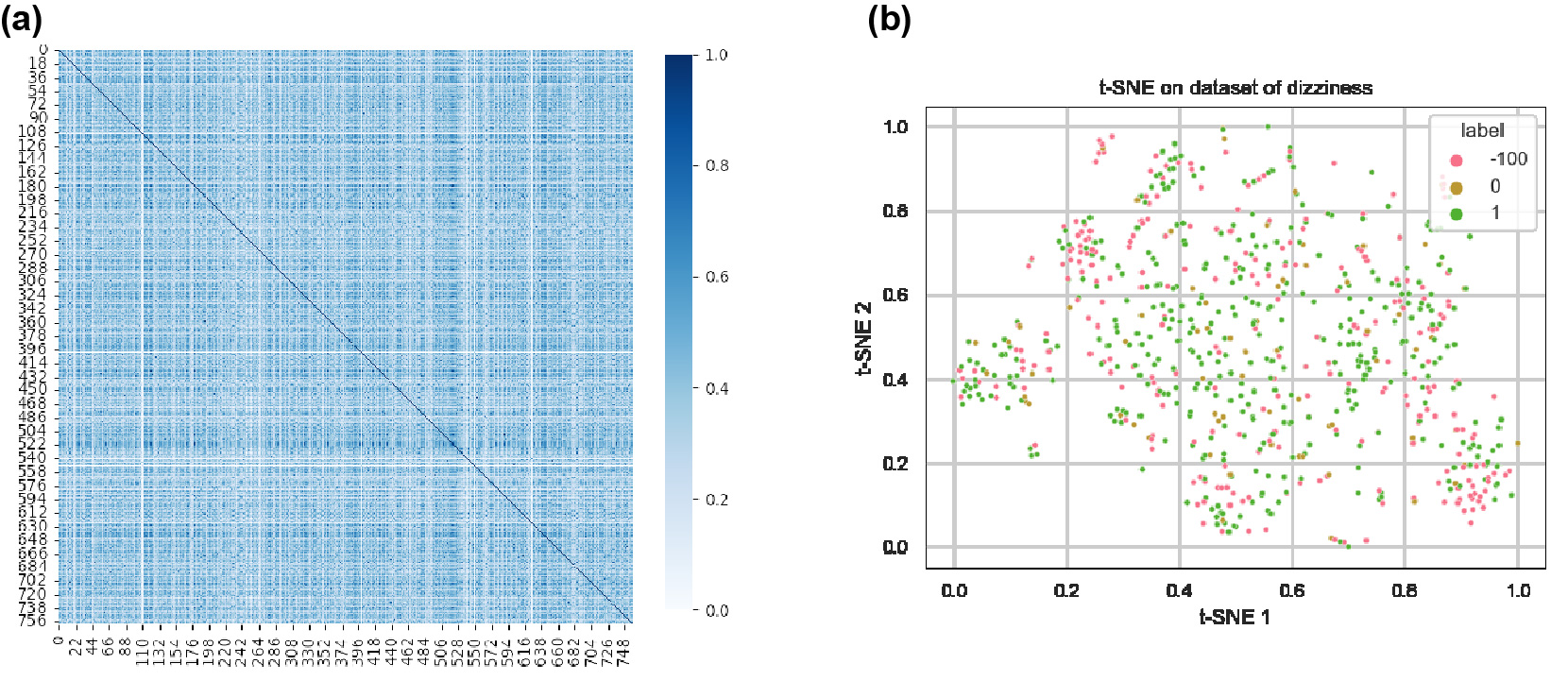 Analysis of the dataset of 759 FDA-approved drugs. (a) The Tanimoto similarity index indicates that the chemical compounds exhibit significant diversity, covering a broad range of chemical space. This demonstrates the structural variability within the dataset. (b) The representative t-Distributed Stochastic Neighbor Embedding (t-SNE) analysis result reveals that conventional dimensionality reduction methods fail to distinguish drugs with observed AEs from those without AEs based solely on chemical structure, underscoring the necessity for advanced modeling techniques like machine learning for AE prediction. Labels in (b) denote frequency of the specified AE: 1 – frequent, 0 – infrequent, -100 – missing values.
Analysis of the dataset of 759 FDA-approved drugs. (a) The Tanimoto similarity index indicates that the chemical compounds exhibit significant diversity, covering a broad range of chemical space. This demonstrates the structural variability within the dataset. (b) The representative t-Distributed Stochastic Neighbor Embedding (t-SNE) analysis result reveals that conventional dimensionality reduction methods fail to distinguish drugs with observed AEs from those without AEs based solely on chemical structure, underscoring the necessity for advanced modeling techniques like machine learning for AE prediction. Labels in (b) denote frequency of the specified AE: 1 – frequent, 0 – infrequent, -100 – missing values.
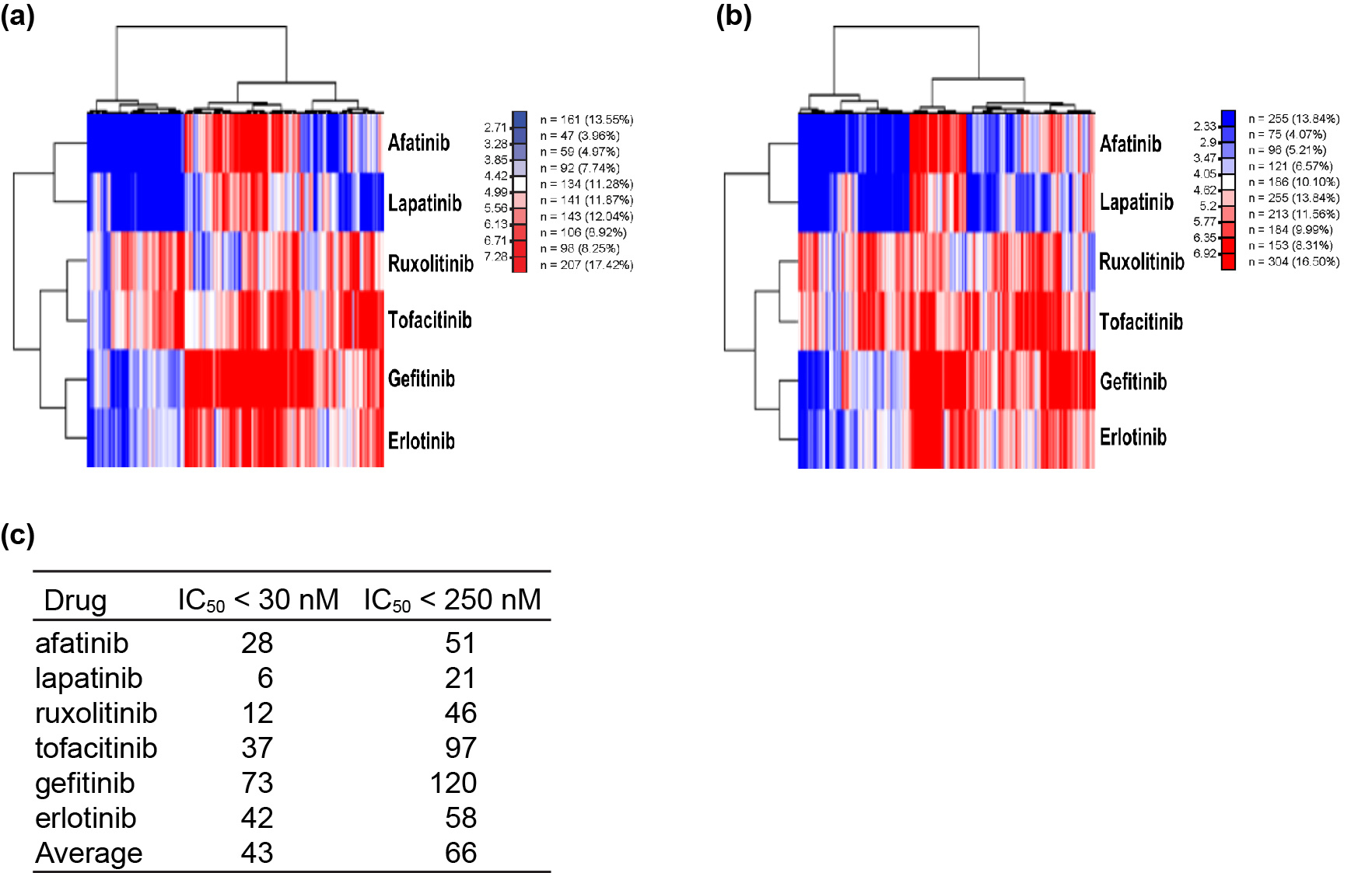 Analysis of predicted on-/off-targets of the six FDA-approved SMKI drugs. Heatmaps of predicted pIC50 values of the SMKIs’ (a) primary targets (IC50 < 30 nM) and (b) primary/unintended targets (IC50 < 250 nM). (c) Summary of the number of predicted on-/off-targets for the six SMKIs.
Analysis of predicted on-/off-targets of the six FDA-approved SMKI drugs. Heatmaps of predicted pIC50 values of the SMKIs’ (a) primary targets (IC50 < 30 nM) and (b) primary/unintended targets (IC50 < 250 nM). (c) Summary of the number of predicted on-/off-targets for the six SMKIs.
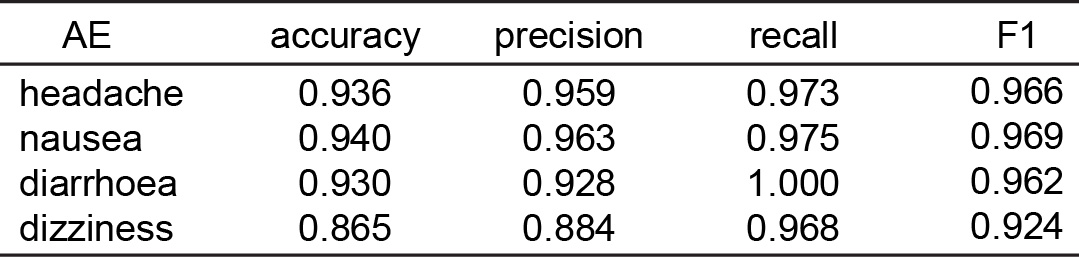 Performance for the AEs prediction
Performance for the AEs prediction
Methods: A collection of 759 FDA-approved drugs with known AEs was adopted directly from Galeano et al.[1]. Protein target expression levels across various tissues were sourced from the Human Protein Atlas[2]. We first adapted the reported ConPLex ML model[3] to predict the drugs’ binding affinities (pIC50) for their intended targets and off-targets among 477 protein targets. Contrastive learning was incorporated to refine the accuracy of the predicted drug-target binding affinities. Subsequently, we developed a multi-layer perceptron (MLP) structure to predict clinical AEs incorporating these predicted drug-target interactions along with the expression profiles of on-/off-targets across various tissues. The model training involved a thorough split of data into training, validation, and testing sets with a ratio of 0.68:0.17:0.15 and utilized weighted binary cross-entropy to address label imbalance within the dataset. The AdamW optimizer (learning rate initialization of 10−5 and weight decay of 10−5) and early stopping techniques were leveraged during model training to optimize performance and mitigate overfitting, respectively.
Results: The utilized dataset of 759 FDA-approved drugs displayed a broad spectrum of structural diversity (Figure 1a), underscoring the versatility of our model in predicting AEs for compounds with a wide range of structural variations. However, traditional dimensionality reduction techniques (Figure 1b) did not effectively segregate compounds based on their associated AEs, emphasizing the multifaceted nature of drug behavior and the need for more advanced modeling approaches to capture these intricate relationships. The adapted ConPLex model predicted drug-target interactions for both on- and off-target effects, as shown by the correct co-localization of the six small molecule kinase inhibitors with their respective kinases (Figure 2). The further constructed MLP model demonstrated good predictive capability of clinical AEs such as headache, nausea, diarrhea, and dizziness, yielding high metrics for accuracy (exceeding 0.86), precision (exceeding 0.88), recall (exceeding 0.96), and F1-scores (exceeding 0.92) (Table 1).
Conclusion: This study presents an advanced machine learning model that integrates drug structure data, on- and off-target binding interactions, and tissue-specific target expression profiles for the prediction of clinical AEs. Our approach provides a new insight into the mechanistic understanding of in vivo drug toxicity in relationship with drug on-/off-target interactions. The coupled ML models, once validated with larger datasets, may offer advantages to directly predict clinical AEs using in vitro/ex vivo and preclinical data, which will help to reduce drug development failure due to clinical toxicity.
References: [1] Galeano D, Li S, Gerstein M, Paccanaro A. Predicting the frequencies of drug side effects. Nature Communications. 2020 Sep 11;11(1):4575.
[2] Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, Olsson I. Tissue-based map of the human proteome. Science. 2015 Jan 23;347(6220):1260419.
[3] Singh R, Sledzieski S, Bryson B, Cowen L, Berger B. Contrastive learning in protein language space predicts interactions between drugs and protein targets. Proceedings of the National Academy of Sciences. 2023 Jun 13;120(24):e2220778120.
Acknowledgements: The authors declare no conflict of interest.
 Analysis of the dataset of 759 FDA-approved drugs. (a) The Tanimoto similarity index indicates that the chemical compounds exhibit significant diversity, covering a broad range of chemical space. This demonstrates the structural variability within the dataset. (b) The representative t-Distributed Stochastic Neighbor Embedding (t-SNE) analysis result reveals that conventional dimensionality reduction methods fail to distinguish drugs with observed AEs from those without AEs based solely on chemical structure, underscoring the necessity for advanced modeling techniques like machine learning for AE prediction. Labels in (b) denote frequency of the specified AE: 1 – frequent, 0 – infrequent, -100 – missing values.
Analysis of the dataset of 759 FDA-approved drugs. (a) The Tanimoto similarity index indicates that the chemical compounds exhibit significant diversity, covering a broad range of chemical space. This demonstrates the structural variability within the dataset. (b) The representative t-Distributed Stochastic Neighbor Embedding (t-SNE) analysis result reveals that conventional dimensionality reduction methods fail to distinguish drugs with observed AEs from those without AEs based solely on chemical structure, underscoring the necessity for advanced modeling techniques like machine learning for AE prediction. Labels in (b) denote frequency of the specified AE: 1 – frequent, 0 – infrequent, -100 – missing values.  Analysis of predicted on-/off-targets of the six FDA-approved SMKI drugs. Heatmaps of predicted pIC50 values of the SMKIs’ (a) primary targets (IC50 < 30 nM) and (b) primary/unintended targets (IC50 < 250 nM). (c) Summary of the number of predicted on-/off-targets for the six SMKIs.
Analysis of predicted on-/off-targets of the six FDA-approved SMKI drugs. Heatmaps of predicted pIC50 values of the SMKIs’ (a) primary targets (IC50 < 30 nM) and (b) primary/unintended targets (IC50 < 250 nM). (c) Summary of the number of predicted on-/off-targets for the six SMKIs. Performance for the AEs prediction
Performance for the AEs prediction