Preclinical, Clinical, and Translational Sciences
(T1530-02-07) Establishing a Dissolution Safe Space for Ibuprofen as a Model BCS Class IIa Drug Using Product Particle Size Distribution (P-PSD) Approaches to Support Biowaiver Consideration
Tuesday, October 22, 2024
3:30 PM - 4:30 PM MT
- YX
Yunming Xu, Pharm.D.
Biopharmaceutics Reviewer
US Food and Drug Administration
Silver Spring, Maryland, United States - MA
Md Haider Ali, Ph.D.
ORISE Fellow
US Food and Drug Administration
Silver Spring, Maryland, United States - IP
Ishwor Poudel, Ph.D.
ORISE Fellow
US Food and Drug Administration
St Louis, Missouri, United States - PC
Parnali Chatterjee, Ph.D.
Pharmacologist
US Food and Drug Administration
Silver Spring, Maryland, United States - ZG
Zongming Gao, Ph.D.
Research Chemist
US Food and Drug Administration
St Louis, Missouri, United States - BR
Bhagwant Rege, Ph.D.
Division Director
US Food and Drug Administration
Silver Spring, Maryland, United States
Presenting Author(s)
Main Author(s)
Co-Author(s)
Purpose: The Biopharmaceutics Classification System (BCS) categorizes drug substances based on their permeability, solubility, and dissolution characteristics (1). Presently, BCS Class I (high solubility and permeability) and Class III (high solubility, low permeability) drugs can obtain waivers for in vivo bioequivalence studies (biowaivers) according to the ICH M9 guidelines. BCS Class II drugs are characterized by high permeability but low solubility. However, BCS Class II acidic (IIa) drugs exhibit behavior similar to BCS Class I drugs at the intestinal pH range of 6.5-7.0, despite their low solubility in acidic (gastric) environments. This project aims to explore a potential biowaiver for BCS Class IIa drugs by establishing a dissolution "safe space" using Product Particle Size Distribution (P-PSD) classic and hydrodynamics (HD) approaches.
Methods: The baseline physiologically based biopharmaceutics model (PBBM) was developed using GastroPlus 9.9. To characterize in vitro dissolution, the P-PSD classic and the P-PSD hydrodynamics (HD) models were employed. P-PSDs were extracted from dissolution data obtained using USP apparatus II, with 900 mL of pH 5.6 phosphate buffer at 50 rpm and 37°C. The P-PSDs were then validated with measured dissolution profile data at pH 4.5 and pH 7.2, as pH 1.2 did not provide meaningful information for validation (7). The goodness-of-fit for the P-PSD models was assessed using Average Fold Error (AFE) and Absolute Average Fold Error (AAFE) (5). The absorption model was configured with default settings for a healthy patient in the fasting state; however, the default bulk pH values for each compartment were modified to reflect the surface pH (2). A previously verified compartmental model was used to describe the drug disposition. The baseline model was validated against clinical data for the same drug product (4). For model application, dissolution profiles were proposed for virtual batches and the corresponding P-PSDs were extracted using both classic and HD approaches. The P-PSDs were subsequently used as input in the PBBM, and then the model was used to predict the relative exposure of ibuprofen for the virtual batches in comparison to the baseline model. Virtual bioequivalence (VBE) trials were performed to identify a dissolution “safe space” (3,6) wherein products exhibiting release profiles within the identified boundaries would be bioequivalent to the reference (i.e., baseline model).
Results: The P-PSDs were extracted using one bin (75 μm at 100%) for the classic model and two bins (75 μm at 26% and 230 μm at 74%) for the hydrodynamics model. Both models reasonably predicted the dissolution profiles at pH 4.5 and 7.2, as shown in Figure 1 and Table 1. When the P-PSDs were integrated into the PBBM via the Johnson model, both approaches provided good estimations of Cmax, AUCt, and AUCi. For the dissolution safe space, both approaches indicated that the upper bound of the dissolution safe space could be close to instant dissolution, as P-PSDs set to 1 μm at 100% still demonstrated bioequivalence (BE) to the baseline. Both models also suggested that even at very slow drug release, represented by one bin with 300 μm at 100% for the classic model, or two bins with 150 μm at 26% and 460 μm at 74% for the HD model, would still indicate BE, resulting in an extremely wide dissolution safe space for both approaches, as shown in Figure 2.
Conclusion: Using a pH 5.6 medium allows for discrimination between different formulations of this drug. The results show that there is no significant difference between the classic and hydrodynamics models, indicating that hydrodynamics is not a significant contributor to in vitro dissolution in this case. The dissolution safe space for both models was extremely wide, suggesting that dissolution is not the rate limiting step with respect to exposure of ibuprofen or the pharmacokinetic performance of this drug. This wide dissolution safe space can be a potential justification for a biowaiver, simplifying the bioequivalence assessment process for BCS Class IIa drugs. However, some limitations were identified: the contribution of bile salts was not well captured, as the in vitro dissolution tests in biorelevant media are ongoing; the validation data were limited, which could be strengthened with more in-depth studies on other formulations of the same drug; the bio-batch used for clinical studies was not the same as the batch used in the in vitro dissolution tests, which may introduce batch-to-batch variations to the model, and the in vitro dissolution profile data demonstrated incomplete release in pH 5.6 medium, which could be addressed by a longer collection time or an additional phase with higher agitation speed. Further research is ongoing to determine whether these findings are drug-specific or if the wide dissolution safe space for ibuprofen is applicable and generalizable to all BCS class IIa drug products. If the findings can be confirmed for other drug products, this may present a case for the expansion of BCS-based biowaivers to include BCS Class IIa drug products.
References: 1. Amidon GL, Lennernas H, Shah VP, Crison JR. A theoretical basis for a biopharmaceutic drug classification: the correlation of in vitro drug product dissolution and in vivo bioavailability. Pharm Res. 1995;12(3):413-20. doi: 10.1023/a:1016212804288. PubMed PMID: 7617530.
2. Hens B, Seegobin N, Bermejo M, Tsume Y, Clear N, McAllister M, et al. Dissolution Challenges Associated with the Surface pH of Drug Particles: Integration into Mechanistic Oral Absorption Modeling. AAPS J. 2022;24(1):17. Epub 20220104. doi: 10.1208/s12248-021-00663-0. PubMed PMID: 34982285.
3. Kourentas A, Gajewska M, Lin W, Dhareshwar SS, Steib-Lauer C, Kulkarni S, et al. Establishing the Safe Space via Physiologically Based Biopharmaceutics Modeling. Case Study: Fevipiprant/QAW039. AAPS J. 2023;25(1):25. Epub 20230214. doi: 10.1208/s12248-023-00787-5. PubMed PMID: 36788163.
4. Pavliv L, Voss B, Rock A. Pharmacokinetics, safety, and tolerability of a rapid infusion of i.v. ibuprofen in healthy adults. Am J Health Syst Pharm. 2011;68(1):47-51. doi: 10.2146/ajhp100120. PubMed PMID: 21164065.
5. Pepin X, Goetschy M, Abrahmsen-Alami S. Mechanistic Models for USP2 Dissolution Apparatus, Including Fluid Hydrodynamics and Sedimentation. J Pharm Sci. 2022;111(1):185-96. Epub 20211016. doi: 10.1016/j.xphs.2021.10.006. PubMed PMID: 34666045.
6. Pepin XJ, Flanagan TR, Holt DJ, Eidelman A, Treacy D, Rowlings CE. Justification of Drug Product Dissolution Rate and Drug Substance Particle Size Specifications Based on Absorption PBPK Modeling for Lesinurad Immediate Release Tablets. Mol Pharm. 2016;13(9):3256-69. Epub 20160727. doi: 10.1021/acs.molpharmaceut.6b00497. PubMed PMID: 27438964.
7. Poudel I, Doole FT, Chatterjee P, Ali MH, Xu Y, Rege B, et al. Development of Biorelevant Dissolution Methods for Potential Risk-Based Biowaivers of BCS Class IIa compounds: Impact of pH, Buffer Capacity and Hydrodynamics of Dissolution Media. AAPS 2024 PharmSci 360 Salt Lake City, UT, USA2024.
Acknowledgements: The present work reflects the view of the author(s) and should not be construed to represent US-FDA’s views or policies.
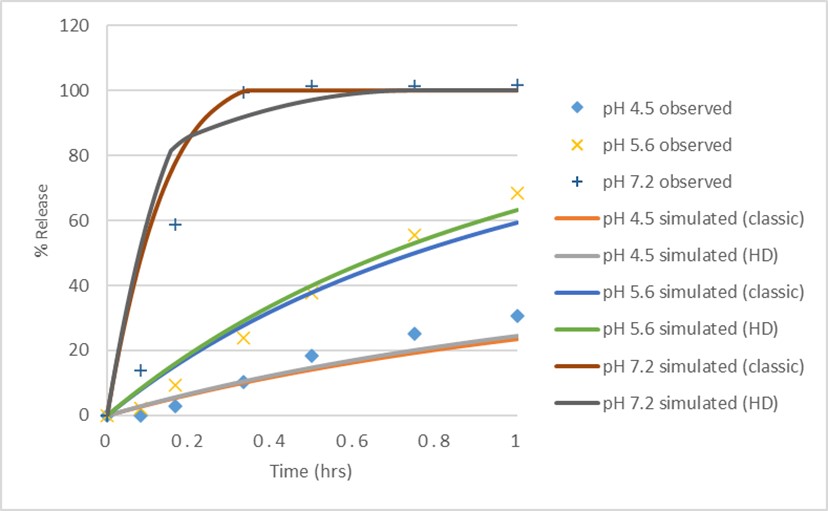 Figure 1: Simulated and observed dissolution profiles using P-PSD classic vs P-PSD HD models in pH 5.6 medium, pH 4.5 medium, and pH 7.2 medium. P-PSD extracted from pH 5.6 dissolution data and prediction performance of P-PSD simulated curve assessed against pH 4.5 and pH 7.2 dissolution data.
Figure 1: Simulated and observed dissolution profiles using P-PSD classic vs P-PSD HD models in pH 5.6 medium, pH 4.5 medium, and pH 7.2 medium. P-PSD extracted from pH 5.6 dissolution data and prediction performance of P-PSD simulated curve assessed against pH 4.5 and pH 7.2 dissolution data.
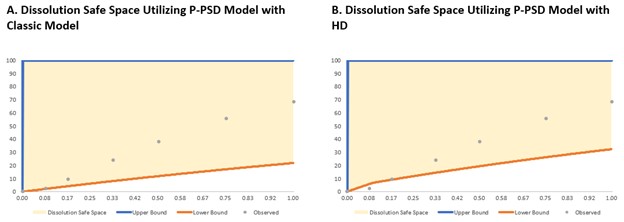 Figure 2: Optimization of dissolution safe space utilizing P-PSD model. For the classic model, as shown at Figure 2A, the upper bound of dissolution safe space was created by setting P-PSD to 1 µm at 100% for an instant complete release and the lower bound of dissolution safe space was created by setting P-PSD to 300 µm at 100% for very slow drug release. For the HD Model shown in Figure 2B, the P-PSD was set to 1 μm at 100% for an instant complete release to generate the upper bound of dissolution safe space and the lower bound of the dissolution safe space was modeled by two bins with 150 μm at 26% and 460 μm at 74%.
Figure 2: Optimization of dissolution safe space utilizing P-PSD model. For the classic model, as shown at Figure 2A, the upper bound of dissolution safe space was created by setting P-PSD to 1 µm at 100% for an instant complete release and the lower bound of dissolution safe space was created by setting P-PSD to 300 µm at 100% for very slow drug release. For the HD Model shown in Figure 2B, the P-PSD was set to 1 μm at 100% for an instant complete release to generate the upper bound of dissolution safe space and the lower bound of the dissolution safe space was modeled by two bins with 150 μm at 26% and 460 μm at 74%.
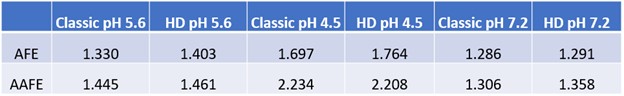 Table 1: AFE and AAFE for evaluating Goodness of Fit for P-PSD classic and P-PSD HD simulated dissolution curves vs observed in-vitro dissolution profiles.
Table 1: AFE and AAFE for evaluating Goodness of Fit for P-PSD classic and P-PSD HD simulated dissolution curves vs observed in-vitro dissolution profiles.
Methods: The baseline physiologically based biopharmaceutics model (PBBM) was developed using GastroPlus 9.9. To characterize in vitro dissolution, the P-PSD classic and the P-PSD hydrodynamics (HD) models were employed. P-PSDs were extracted from dissolution data obtained using USP apparatus II, with 900 mL of pH 5.6 phosphate buffer at 50 rpm and 37°C. The P-PSDs were then validated with measured dissolution profile data at pH 4.5 and pH 7.2, as pH 1.2 did not provide meaningful information for validation (7). The goodness-of-fit for the P-PSD models was assessed using Average Fold Error (AFE) and Absolute Average Fold Error (AAFE) (5). The absorption model was configured with default settings for a healthy patient in the fasting state; however, the default bulk pH values for each compartment were modified to reflect the surface pH (2). A previously verified compartmental model was used to describe the drug disposition. The baseline model was validated against clinical data for the same drug product (4). For model application, dissolution profiles were proposed for virtual batches and the corresponding P-PSDs were extracted using both classic and HD approaches. The P-PSDs were subsequently used as input in the PBBM, and then the model was used to predict the relative exposure of ibuprofen for the virtual batches in comparison to the baseline model. Virtual bioequivalence (VBE) trials were performed to identify a dissolution “safe space” (3,6) wherein products exhibiting release profiles within the identified boundaries would be bioequivalent to the reference (i.e., baseline model).
Results: The P-PSDs were extracted using one bin (75 μm at 100%) for the classic model and two bins (75 μm at 26% and 230 μm at 74%) for the hydrodynamics model. Both models reasonably predicted the dissolution profiles at pH 4.5 and 7.2, as shown in Figure 1 and Table 1. When the P-PSDs were integrated into the PBBM via the Johnson model, both approaches provided good estimations of Cmax, AUCt, and AUCi. For the dissolution safe space, both approaches indicated that the upper bound of the dissolution safe space could be close to instant dissolution, as P-PSDs set to 1 μm at 100% still demonstrated bioequivalence (BE) to the baseline. Both models also suggested that even at very slow drug release, represented by one bin with 300 μm at 100% for the classic model, or two bins with 150 μm at 26% and 460 μm at 74% for the HD model, would still indicate BE, resulting in an extremely wide dissolution safe space for both approaches, as shown in Figure 2.
Conclusion: Using a pH 5.6 medium allows for discrimination between different formulations of this drug. The results show that there is no significant difference between the classic and hydrodynamics models, indicating that hydrodynamics is not a significant contributor to in vitro dissolution in this case. The dissolution safe space for both models was extremely wide, suggesting that dissolution is not the rate limiting step with respect to exposure of ibuprofen or the pharmacokinetic performance of this drug. This wide dissolution safe space can be a potential justification for a biowaiver, simplifying the bioequivalence assessment process for BCS Class IIa drugs. However, some limitations were identified: the contribution of bile salts was not well captured, as the in vitro dissolution tests in biorelevant media are ongoing; the validation data were limited, which could be strengthened with more in-depth studies on other formulations of the same drug; the bio-batch used for clinical studies was not the same as the batch used in the in vitro dissolution tests, which may introduce batch-to-batch variations to the model, and the in vitro dissolution profile data demonstrated incomplete release in pH 5.6 medium, which could be addressed by a longer collection time or an additional phase with higher agitation speed. Further research is ongoing to determine whether these findings are drug-specific or if the wide dissolution safe space for ibuprofen is applicable and generalizable to all BCS class IIa drug products. If the findings can be confirmed for other drug products, this may present a case for the expansion of BCS-based biowaivers to include BCS Class IIa drug products.
References: 1. Amidon GL, Lennernas H, Shah VP, Crison JR. A theoretical basis for a biopharmaceutic drug classification: the correlation of in vitro drug product dissolution and in vivo bioavailability. Pharm Res. 1995;12(3):413-20. doi: 10.1023/a:1016212804288. PubMed PMID: 7617530.
2. Hens B, Seegobin N, Bermejo M, Tsume Y, Clear N, McAllister M, et al. Dissolution Challenges Associated with the Surface pH of Drug Particles: Integration into Mechanistic Oral Absorption Modeling. AAPS J. 2022;24(1):17. Epub 20220104. doi: 10.1208/s12248-021-00663-0. PubMed PMID: 34982285.
3. Kourentas A, Gajewska M, Lin W, Dhareshwar SS, Steib-Lauer C, Kulkarni S, et al. Establishing the Safe Space via Physiologically Based Biopharmaceutics Modeling. Case Study: Fevipiprant/QAW039. AAPS J. 2023;25(1):25. Epub 20230214. doi: 10.1208/s12248-023-00787-5. PubMed PMID: 36788163.
4. Pavliv L, Voss B, Rock A. Pharmacokinetics, safety, and tolerability of a rapid infusion of i.v. ibuprofen in healthy adults. Am J Health Syst Pharm. 2011;68(1):47-51. doi: 10.2146/ajhp100120. PubMed PMID: 21164065.
5. Pepin X, Goetschy M, Abrahmsen-Alami S. Mechanistic Models for USP2 Dissolution Apparatus, Including Fluid Hydrodynamics and Sedimentation. J Pharm Sci. 2022;111(1):185-96. Epub 20211016. doi: 10.1016/j.xphs.2021.10.006. PubMed PMID: 34666045.
6. Pepin XJ, Flanagan TR, Holt DJ, Eidelman A, Treacy D, Rowlings CE. Justification of Drug Product Dissolution Rate and Drug Substance Particle Size Specifications Based on Absorption PBPK Modeling for Lesinurad Immediate Release Tablets. Mol Pharm. 2016;13(9):3256-69. Epub 20160727. doi: 10.1021/acs.molpharmaceut.6b00497. PubMed PMID: 27438964.
7. Poudel I, Doole FT, Chatterjee P, Ali MH, Xu Y, Rege B, et al. Development of Biorelevant Dissolution Methods for Potential Risk-Based Biowaivers of BCS Class IIa compounds: Impact of pH, Buffer Capacity and Hydrodynamics of Dissolution Media. AAPS 2024 PharmSci 360 Salt Lake City, UT, USA2024.
Acknowledgements: The present work reflects the view of the author(s) and should not be construed to represent US-FDA’s views or policies.
 Figure 1: Simulated and observed dissolution profiles using P-PSD classic vs P-PSD HD models in pH 5.6 medium, pH 4.5 medium, and pH 7.2 medium. P-PSD extracted from pH 5.6 dissolution data and prediction performance of P-PSD simulated curve assessed against pH 4.5 and pH 7.2 dissolution data.
Figure 1: Simulated and observed dissolution profiles using P-PSD classic vs P-PSD HD models in pH 5.6 medium, pH 4.5 medium, and pH 7.2 medium. P-PSD extracted from pH 5.6 dissolution data and prediction performance of P-PSD simulated curve assessed against pH 4.5 and pH 7.2 dissolution data.  Figure 2: Optimization of dissolution safe space utilizing P-PSD model. For the classic model, as shown at Figure 2A, the upper bound of dissolution safe space was created by setting P-PSD to 1 µm at 100% for an instant complete release and the lower bound of dissolution safe space was created by setting P-PSD to 300 µm at 100% for very slow drug release. For the HD Model shown in Figure 2B, the P-PSD was set to 1 μm at 100% for an instant complete release to generate the upper bound of dissolution safe space and the lower bound of the dissolution safe space was modeled by two bins with 150 μm at 26% and 460 μm at 74%.
Figure 2: Optimization of dissolution safe space utilizing P-PSD model. For the classic model, as shown at Figure 2A, the upper bound of dissolution safe space was created by setting P-PSD to 1 µm at 100% for an instant complete release and the lower bound of dissolution safe space was created by setting P-PSD to 300 µm at 100% for very slow drug release. For the HD Model shown in Figure 2B, the P-PSD was set to 1 μm at 100% for an instant complete release to generate the upper bound of dissolution safe space and the lower bound of the dissolution safe space was modeled by two bins with 150 μm at 26% and 460 μm at 74%. Table 1: AFE and AAFE for evaluating Goodness of Fit for P-PSD classic and P-PSD HD simulated dissolution curves vs observed in-vitro dissolution profiles.
Table 1: AFE and AAFE for evaluating Goodness of Fit for P-PSD classic and P-PSD HD simulated dissolution curves vs observed in-vitro dissolution profiles.